This page provides the step-by-step instructions to apply a multi-temporal analysis of the MODIS-based Vegetation Condition Index (VCI) for large areas using R. Depending on the extent of the study area and the hardware capacities of the computer, the user might experiece difficulties applying the original R script including a cloud mask. We therefore provide an additional R script to be able the compute the VCI and monitor the vegetation conditions for large areas (>200.000km2).
For this Recommended Practice for drought monitoring we use the Enhanced Vegetation Index (EVI) based on MODIS data at 250m spatial resolution as input for generating the Vegetation Condition Index (VCI). The only MODIS product providing EVI data is MOD13Q1 (on Terra satellite) and MYD13Q1 (on Aqua Satellite). MODIS data is for example made available on the Application for Extracting and Exploring Analysis Ready Samples (AppEEARS) by USGS.
If the Normalized Difference Vegetation Index (NDVI) is preferred for calculating the VCI, the EVI can easily be replaced with the NDVI as this product is also available for download at AppEEARS. Given this case, the caption of the resulting jpg maps should be adjusted. However, in certain circumstances, the VCI based on the NDVI does not perform as well as based on the EVI. See the "In Detail" page for more information.
Data and software required for this Recommended Practice are:
- Shapefile of area of interest (https://gadm.org/country)
- EVI and Pixel Reliability data provided by MOD13Q1 (see next section)
- R and R-Studio

If not yet installed, the R software and R-Studio can be downloaded here. To follow a step by step instruction on how to download and install R and R-Studio, please click here.
Content
Step 0: Download MODIS data
Step 1: Download R code
Step 2: Adjust the script
Step 3: Run the R code
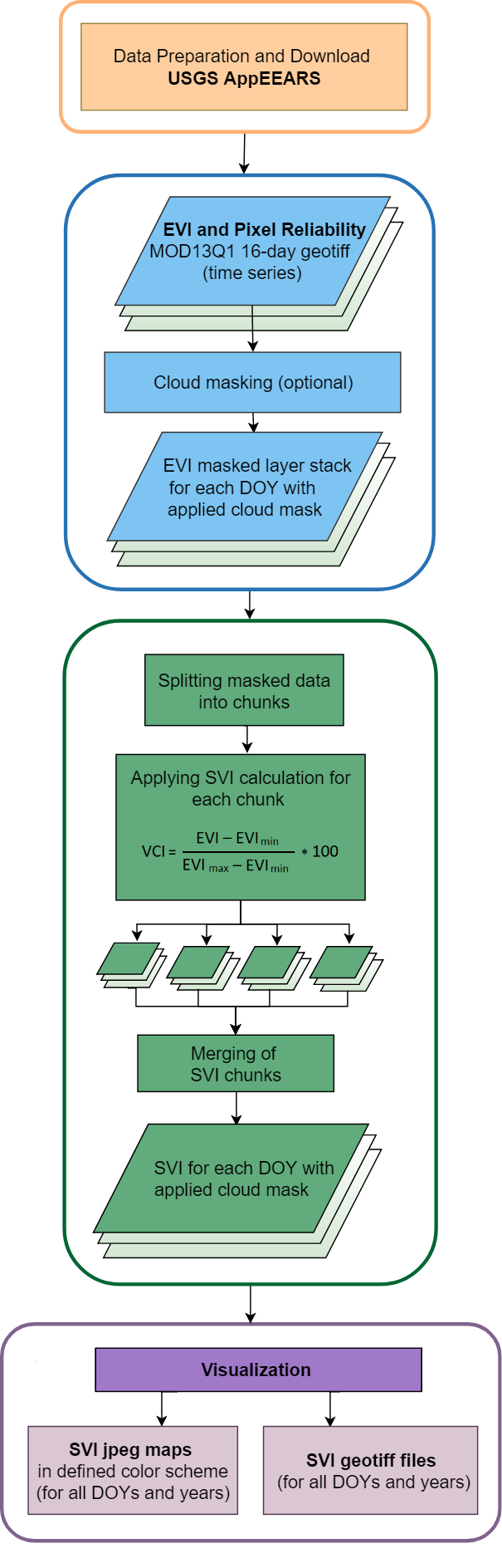
Flowchart of the R code including the cloud mask and chunking
Step 0 Download MODIS data from AppEEARS
Access the AppEEARS website and create a user account (free) in case you do not already have one (https://lpdaacsvc.cr.usgs.gov/appeears/). Click on Extract and Area Sample and start a new request.
- Enter a request name
- Define your region of interest by specifying a ESRI shapefile (zip) or drawing a polygon . Country shapefiles can be downloaded at https://gadm.org/country. Note: The boundaries and names of the shapefiles do not imply official endorsement or acceptance by the United Nations.
- Define the time period of your data: January 2000 to today.
- Select the product: MOD13Q1 and the layers of interest: EVI (or NDVI) and pixel_reliability
- Define the output as geotiff with geographic Projection
- Click on Submit

Click on Explore to check the status of the request. Click on the Download symbol once the status says Done.
Select "All" and save the "download list". The data does not need to be downloaded at this point as it will later be downloaded automatically in R using the download-list. The files can of course also be downloaded manually from AppEEARS (the data then should be stored at the datapath). However, the advantage of using the download-list is to run the data download and data calculation in one go.
Contrary to the data preparation for the original R script including a cloud mask, the data must not be sorted according to the DOY for this R script. Consequently, if you first used the original R script but then had to switch to this R script due to the size of the area, please sort all EVI and Pixel Reliability data into your main directory (datapath, see below). You can avoid moving the files manually by using "command promt" (use "terminal" for Linux or Mac OS).
Note: After selecting "All", the data size of the EVI and Pixel Reliablity data (including the VI quality) will be displayed. This information should be taken into consideration especially for large areas to ensure that the datapath used for the calculation has free space available accordingly (for all products and temporary files). However, the VI quality (the Vegetation Index (VI) quality provides similar information as the pixel reliability) will be excluded in the automatical download within R.
Step 1 Download R Code
Download the R Code here (right click and choose "save as"). The R code is tailored to MOD13Q1 tif data.
Step 2 Adjust the script
Open R Studio ant the saved R script (file ->Open File). The script needs small adjustments to fit your data. Note hereby: R uses slashes ("/") for file names and directories and not backslashed ("\").
- In line 5: dataPath<-"C:/Users/Desktop/Peru_VCI" --- Enter the path to the folder where you have enough free space to store your MODIS data, the temporary files and the resulting products. It is recommended to use an external hard drive to store your input and output data if possible
- In line 8: downloadList<-"C:/Users/Desktop/Peru_VCI/peru-download-list.txt" --- enter the path to your download list inlcuding the ".txt" extension
- In line 14-15: aUsername <- "USER", aPassword <- "PASSWORD" --- enter your appEEARS/Earthdata username and password here. No account? Register here: https://urs.earthdata.nasa.gov/home
- optional: In line 11: cloudmask<-1 --- replace "1" with "0" if you do not want to apply MODIS pixel reliability data for masking clouds
- optional: In line 248-251: plot(Mos, zlim=c(0,100), col=my_palette(101), main=paste("VCI"," (EVI)"," sample ",DOYs[d]," ",YEARs[y],sep="")) --- Adjust the file naming according to the data you are processing. Especially replace "sample" in line 224 with your study area. Also, if you derived the SVI from the NDVI, change (EV) to (NDVI)
Step 3 Run the script
Install the raster, curl, rgdal and zoo packages by highlighting the respective text in the code and clicking on "Run" or pressing Ctrl+Enter.
Now, run the remaining code by highlighting it and pressing Ctrl+enter. Depending on the size of your data, it will take quite a while (up to several hours) for the script to run - so be patient.
View the Code
You can access the code on https://rpubs.com/
Results
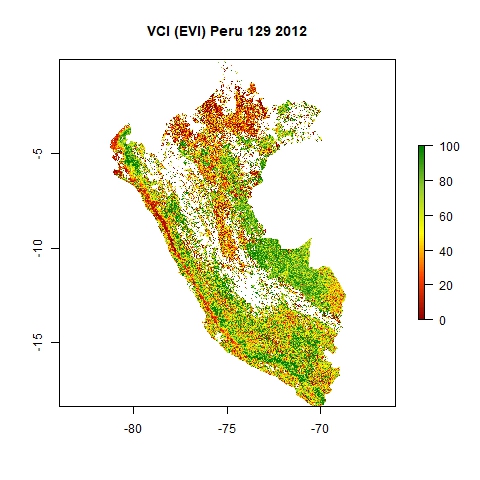
The results will be stored in the folders "VCIjpg" (as jpg) and "VCI" (as GeoTiff) within your main directory. The maps are named "DOY_YYYY". See below an example of a resulting jpg map from DOY 129 in 2012, the 8 May 2012.